FISH
FISH workflow
Fluorescence In Situ Hybridization
Click for more details
The detection of whole-bacterial cells via the labelling of specific nucleic acids with fluorescently labelled oligonucleotide probes is called fluorescence in situ hybridization (FISH). FISH requires no cultivation and cells can be fixed before analysis. Compared to conventional cultivation techniques this method offers distinct advantages:
FISH allows the detection of one to three orders of magnitude more bacterial cells in samples. Even when using optimal media and growth conditions, generally less than 1-10% of the bacteria present in life samples will develop into detectable colonies.
FISH allows the study of the actual composition of the microbial community. When using cultural methods, over-representation as well as under-representation of populations can occur due to selectivity of the applied cultivation conditions.
FISH allows the in situ localisation and the study of spatial organisation of bacterial cells as they occur in their natural habitat. Unlike immunological methods for detection, FISH is not dependent on extracellular macromolecules that may only be expressed under certain cultivation conditions, but that are absent in other situations. FISH results are always definitive.
For FISH, cells need not be alive. Samples can easily be fixed and collected for later analysis. The intensity of the fluorescence is a direct measure for the activity of the cells. Inactive cells can be recognised by their low intensity fluorescence.
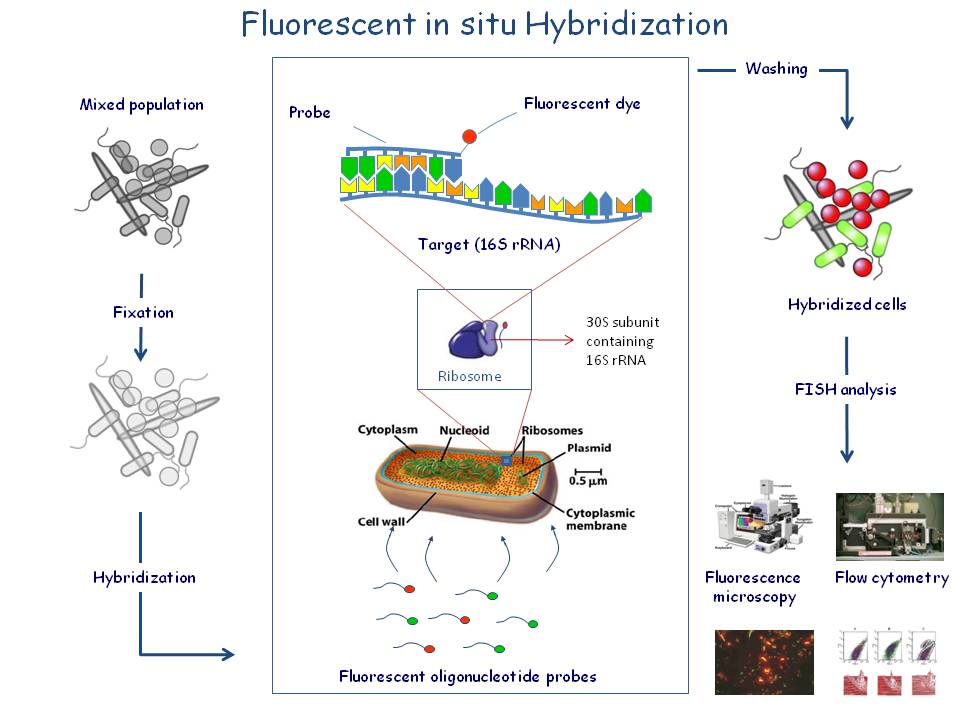
Bacterial FISH probes are often primers for the 16s rRNA molecule. This molecule exists in multiple copies in the cell (up to 10E4-10E5) and is an excellent target for fluorescently labelled oligonucleotide probes which are directed against regions on the rRNA molecule specific for a bacterial group, genus or species. The small probes (16-20 nucleotides) will penetrate the bacterial cell wall and hybridise with the complementary target sequence. Evaluation of the test result is done by epifluorescence microscopy or flow cytometry.
FISH is a very powerful tool to study microbial population structures and dynamics. The method requires no cultivation and cells can be fixed before analysis. FISH allows the in situ localisation and the study of spatial organisation of bacterial cells as they occur in their natural habitat. Unlike immunological methods for detection, FISH is not dependent on extracellular macromolecules that may only be expressed under certain cultivation conditions, but that are absent in other situations. When, for example, attempting to monitor the number of intestinal Bifidobacterium spp. over time by using a series of faecal samples, conventional procedures are too labour-intensive to allow for elaborate experimental designs. Moreover, results from cultivation-based enumerations are statistically less accurate.
FISH results are always definitive. For FISH, cells need not be alive. Samples can easily be fixed and collected for later analysis. The intensity of the fluorescence is a direct measure for the activity of the cells. Inactive cells can be recognised by their low intensity fluorescence. These and other advantages have all been combined in the Ribo Technologies FISH Kit.
Several of the Ribo Technologies FISH Kits are especially designed for the rapid detection and accurate enumeration of bacterial species of the human intestinal microflora. The demonstration of the presence and the accurate enumeration of these bacteria may contribute evidence to the effect of prebiotics, probiotics and antibiotics. In contrast to culturing, the detection and quantification of specific bacterial genera or species with the Ribo Technologies FISH Kit is a very rapid, reliable and accurate method to study the population composition of human intestinal microflora in which the presence of certain bacteria is to be enumerated. Furthermore, the Kit is extremely suitable for rapid identification or culture confirmation purposes.
FISH allows the detection of one to three orders of magnitude more bacterial cells in samples. Even when using optimal media and growth conditions, generally less than 1-10% of the bacteria present in life samples will develop into detectable colonies.
FISH allows the study of the actual composition of the microbial community. When using cultural methods, over-representation as well as under-representation of populations can occur due to selectivity of the applied cultivation conditions.
FISH allows the in situ localisation and the study of spatial organisation of bacterial cells as they occur in their natural habitat. Unlike immunological methods for detection, FISH is not dependent on extracellular macromolecules that may only be expressed under certain cultivation conditions, but that are absent in other situations. FISH results are always definitive.
For FISH, cells need not be alive. Samples can easily be fixed and collected for later analysis. The intensity of the fluorescence is a direct measure for the activity of the cells. Inactive cells can be recognised by their low intensity fluorescence.
Bacterial FISH probes are often primers for the 16s rRNA molecule. This molecule exists in multiple copies in the cell (up to 10E4-10E5) and is an excellent target for fluorescently labelled oligonucleotide probes which are directed against regions on the rRNA molecule specific for a bacterial group, genus or species. The small probes (16-20 nucleotides) will penetrate the bacterial cell wall and hybridise with the complementary target sequence. Evaluation of the test result is done by epifluorescence microscopy or flow cytometry.
FISH is a very powerful tool to study microbial population structures and dynamics. The method requires no cultivation and cells can be fixed before analysis. FISH allows the in situ localisation and the study of spatial organisation of bacterial cells as they occur in their natural habitat. Unlike immunological methods for detection, FISH is not dependent on extracellular macromolecules that may only be expressed under certain cultivation conditions, but that are absent in other situations. When, for example, attempting to monitor the number of intestinal Bifidobacterium spp. over time by using a series of faecal samples, conventional procedures are too labour-intensive to allow for elaborate experimental designs. Moreover, results from cultivation-based enumerations are statistically less accurate.
FISH results are always definitive. For FISH, cells need not be alive. Samples can easily be fixed and collected for later analysis. The intensity of the fluorescence is a direct measure for the activity of the cells. Inactive cells can be recognised by their low intensity fluorescence. These and other advantages have all been combined in the Ribo Technologies FISH Kit.
Several of the Ribo Technologies FISH Kits are especially designed for the rapid detection and accurate enumeration of bacterial species of the human intestinal microflora. The demonstration of the presence and the accurate enumeration of these bacteria may contribute evidence to the effect of prebiotics, probiotics and antibiotics. In contrast to culturing, the detection and quantification of specific bacterial genera or species with the Ribo Technologies FISH Kit is a very rapid, reliable and accurate method to study the population composition of human intestinal microflora in which the presence of certain bacteria is to be enumerated. Furthermore, the Kit is extremely suitable for rapid identification or culture confirmation purposes.