T(D)GGE
Gut microflora analysis services of BioVisible
BioVisible offers a unique analysis service for the evaluation of the effects of probiotics and prebiotics on Gut microflora using FISH (fluorescence in situ hybridization), (T)DGGE ((Temperature)Denaturing Gradient Gel Electrophoresis) and/or Real-time PCR.
Please contact us for more information
T(D)GGE Workflow
T(D)GGE Gel Analysis on Fecal samples

Total DNA is isolated from fecal samples. Using universal primers a discriminating region of the 16S rDNA is amplified. The amplified product is applied on a T(D)GGE gel. After the gelelectrophoresis the gel is silver stained and ready for analysis.
T(D)GGE analysis - Profile determination

The gel is scanned and a profile is generated based on band location and intensity.
T(D)GGE analysis - Profile comparison

The profiles are compared and a cluster analysis is performed.
T(D)GGE analysis - Cluster analysis
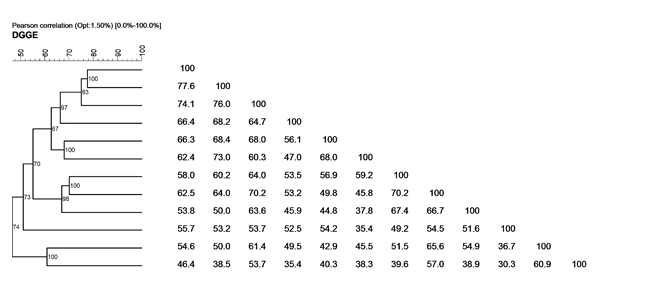
The cluster analysis gives a measure of relatedness of the different fecal profiles. Especially within one subject a T(D)GGE cluster analysis can show the dynamics of the gut microbiota, in time, in relation to diet changes, Pro- Pre-biotica and/or drug administration.
Temperature Gradient Gel Electrophoresis (TGGE) or Denaturing Gradient Gel Electrophoresis (DGGE) is a molecular fingerprinting method that separates polymerase chain reaction (PCR)-generated DNA products. In TGGE there is a temperature gradient across the gel. in DGGE there is a chemical gradient across the gel. TGGE and DGGE are useful for analyzing nucleic acids such as DNA and RNA, and sometimes for proteins.
The polymerase chain reaction of environmental DNA of a single target can generate templates of differing DNA sequence that represent many of the dominant microbial organisms. However, since PCR products from a given reaction are of similar size (bp), conventional separation by agarose gel electrophoresis results only in a single DNA band that is largely non-descriptive. T(D)GGE can overcome this limitation by separating PCR products based on sequence differences that results in differential denaturing characteristics of the DNA.
During T(D)GGE, PCR products encounter increasingly higher temperature or concentrations of chemical denaturant as they migrate through a polyacrylamide gel. Upon reaching a threshold temperature or denaturant concentration, the weaker melting domains of the double-stranded PCR product will begin to denature at which time migration slows dramatically. Differing sequences of DNA (from different bacteria) will denature at different temperature or denaturant concentrations resulting in a pattern of bands. Each band theoretically representing a different bacterial population present in the community.
In essence, DGGE is based on the principle that, in a polyacrylamide matrix, double stranded DNA’s migrate at a higher rate than their (partially) single stranded counterparts. The introduction of a G+C-rich “clamp” to one side of the double stranded DNA will keep the two strands connected to each other even when relatively strong denaturing conditions are met. Due to this GC-clamp, the difference in migration rate between double stranded and single stranded DNA’s is dramatically improved. In fully denatured form, the migration of the DNA fragments is virtually arrested.
The possibility to effectively separate many (>20) different ribosomal RNA genes is further based on the principle that strand separation or denaturation of the dsDNA’s is influenced by the G+C content of the DNA’s and the presence of “melting regions” (i.e., partially A+T-rich sequences at which strand separating is initiated earlier than in the rest of the helix thus retarding the migration of the fragment).
In order to obtain the (partial) ribosomal RNA genes from all the bacteria in a complex mixed population, the genomic DNA extracted from the total population is used as a template in a PCR reaction. Rigorous DNA extraction procedures should be used in order to ensure that genomic DNA is liberated from all bacteria. Specific regions of this mixed rRNA gene pool are amplified by using a PCR primer pair directed towards (universally) conserved gene sequences. Only one of the PCR primers contains the GC-clamp. After gel-electrophoretic separation of these PCR products on a chemical gradient of urea and silver staining of the DNA, a banding pattern emerges that represents the diversity of the ribosomal RNA gene sequences present in the sample. The intensity of the individual bands is a measure for the relative abundance of this sequence in the population. Every band originates - in principle - from one bacterial species, although certain PCR bias towards A+T-rich sequences may occur.
Computer analysis of the DGGE profiles of the samples results in similarity percentages between the individual samples. DGGE is a reproducible form of TGGE, in which the separation of DNA’s is not dependent on a temperature gradient but on a chemical gradient of urea.
DGGE is an exceptional tool to study the bacterial species composition of unknown samples. By inter-sample comparison, dominant shifts in population composition can be monitored without any prior knowledge of that sample. By using DGGE, bacterial population dynamics can be studied in more detail.